| SAG Strain Number: | 104.79 |
| TAXONOMY |
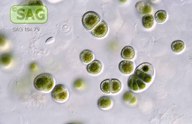
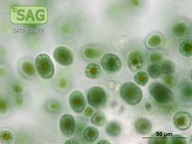
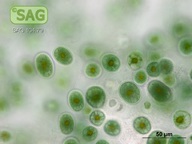
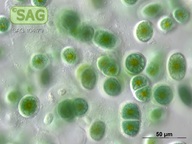
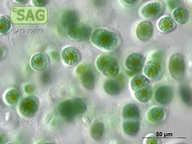
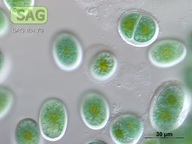
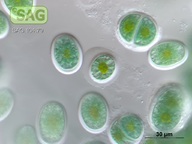
| Genus: | Chroothece |
| Species: | richteriana |
| Taxonomic position: | Rhodophyta - Rhodophyceae |
| Authority: | Hansgirg |
| Variety: | |
| Formerly called: | |
| Authentic: | no |
| Division: | Rhodophyta |
| Class: | Rhodophyceae |
| Common name: | Red Algae - Rhodophytes |
| ORIGIN |
| General habitat: | epilithic (surface of rocks and buildings etc.) |
| Climatic zone: | |
| Continent: | America |
| Country: | USA |
| Locality: | TX, Hamiltons Pool in Travis Co. |
| Lat. / Long. (Precision): | 30.342213 / -98.127229 (500m) View on Google Maps |
| Year: | before 1978 |
| Isolated by: | F.D. Ott |
| Strain number by isolator: | MO 448 |
| Deposition by: | F.D. Ott |
| Deposition date: | 1978 |
| CULTURE INFORMATION |
| Culture medium: | 1/2SWES |
| Axenic: | bacterial or other types of contamination |
| Strain relatives: | probably CCAP 1353/4 |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain |
| |
| PUBLICATIONS |
| Publication: | Aboal M. et al. (2014) Ecology, morphology and physiology of Chroothece richteriana (Rhodophyta, Stylonematophyceae) in the highly calcareous Río Chícamo, south-east Spain. Eur. J. Phycol. 49 (1):83-96 |
| Publication: | Lang I. et al. (2011) Fatty acid profiles and their distribution patterns in microalgae: a comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol 11:124 |
| Publication: | Oliveira, M.C. et al. (2000) Phylogeny of the Bangiophycidae (Rhodophyta) and the secondary endosymbiotic origin of algal plastids. Am. J. Bot. 87 (4):482-492 |
| SEQUENCE INFORMATION |
| Sequence Accession: | OK643923 Chroothece mobilis NADH dehydrogenase subunit 5 (nad5) gene, partial cds; mitochondrial. (753 bp) |
| Sequence Accession: | OK643921 Chroothece mobilis NADH dehydrogenase subunit 4 (nad4) gene, complete cds; mitochondrial. (1485 bp) |
| Sequence Accession: | OK643919 Chroothece mobilis NADH dehydrogenase subunit 2 (nad2) gene, complete cds; mitochondrial. (1437 bp) |
| Sequence Accession: | OK643918 Chroothece mobilis NADH dehydrogenase subunit 1 (nad1) gene, complete cds; mitochondrial. (1014 bp) |
| Sequence Accession: | OK643917 Chroothece mobilis cytochrome c oxidase subunit III (cox3) gene, complete cds; mitochondrial. (798 bp) |
| Sequence Accession: | OK643916 Chroothece mobilis cytochrome c oxidase subunit II (cox2) gene, complete cds; mitochondrial. (765 bp) |
| Sequence Accession: | OK643915 Chroothece mobilis cytochrome c oxidase subunit I (cox1) gene, complete cds; mitochondrial. (1572 bp) |
| Sequence Accession: | OK643914 Chroothece mobilis cytochrome b (cob) gene, complete cds; mitochondrial. (1140 bp) |
| Sequence Accession: | OK643912 Chroothece mobilis ATP synthase F0 subunit 6 (atp6) gene, complete cds; mitochondrial. (747 bp) |
| Sequence Accession: | OK643924 Chroothece mobilis succinate dehydrogenase subunit 2 (sdh2) gene, complete cds; mitochondrial. (843 bp) |
| Sequence Accession: | OK643913 Chroothece mobilis ATP synthase F0 subunit 9 (atp9) gene, complete cds; mitochondrial. (225 bp) |
| Sequence Accession: | OK643920 Chroothece mobilis NADH dehydrogenase subunit 3 (nad3) gene, complete cds; mitochondrial. (312 bp) |
| Sequence Accession: | OK643922 Chroothece mobilis NADH dehydrogenase subunit 4L (nad4L) gene, complete cds; mitochondrial. (285 bp) |
| Sequence Accession: | NC_062379 Chroothece richteriana strain SAG 104.79 chloroplast, complete genome. (207333 bp) |
| Sequence Accession: | MZ680365 Chroothece richteriana strain SAG 104.79 chloroplast, complete genome. (207333 bp) |
| Sequence Accession: | AF170711 Chroothece richteriana strain SAG B 104.79 culture-collection SAG:104.79 16S small subunit ribosomal RNA gene, partial sequence; plastid. (1439 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | yes |