| SAG Strain Number: | 11-32b |
| TAXONOMY |
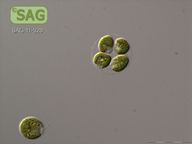
| Genus: | Chlamydomonas |
| Species: | reinhardtii |
| Taxonomic position: | Chlorophyta - Chlorophyceae |
| Authority: | Dangeard |
| Variety: | |
| Formerly called: | |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN |
| General habitat: | soil |
| Climatic zone: | |
| Continent: | America |
| Country: | USA |
| Locality: | MA, soil from potato field near Amherst |
| Lat. / Long. (Precision): | 42.364125 / -72.507706 (10000m) View on Google Maps |
| Year: | 1945 |
| Isolated by: | G. M. Smith |
| Strain number by isolator: | |
| Deposition by: | UTEX |
| Deposition date: | before 1964 |
| CULTURE INFORMATION |
| Culture medium: | K Ag |
| Axenic: | axenic |
| Strain relatives: | CCAP 11/32A; UTEX 90 |
| Agitation resistance: | yes |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; recommended for schools, + mating type, dioecious |
| |
| PUBLICATIONS |
| Publication: | Fragkopoulos A.A. et al. (2025) Metabolic activity controls the emergence of coherent flows in microbial suspensions. Proc. Natl. Acad. Sci. U.S.A. 122 (4):e2413340122 |
| Publication: | Wagner H. et al. (2024) Carbon and energy balance of biotechnological glycolate production from microalgae in a pre-industrial scale flat panel photobioreactor. Biotechnol. Biofuels 17:42 |
| Publication: | Weithoff G. et al. (2024) Weak effect of temperature fluctuations on the invasion of Raphidiopsis raciborskii (Cyanobacteria) in experimental plankton microcosms. J. Phycol. 61:261-266 |
| Publication: | Catalan R.E. et al. (2023) Light-regulated adsorption and desorption of Chlamydomonas cells at surfaces. Soft Matter 19:306-314 |
| Publication: | Schad A. et al. (2023) Optimising biotechnological glycolate production in Chlamydomonas reinhardtii by improving carbon allocation towards the product. Chemical Engineering Journal 459:141432 |
| Publication: | Harth F.M. et al. (2022) Ru/C-Catalyzed Hydrogenation of Aqueous Glycolic Acid from Microalgae – Influence of pH and Biologically Relevant Additives. ChemistryOpen 11:e202200050 |
| Publication: | Schad A. et al. (2022) Crossing and selection of Chlamydomonas reinhardtii strains for biotechnological glycolate production. Appl. Microbiol. Biotechnol. 106:3539-3554 |
| Publication: | Till S. et al. (2022) Motility and self-organization of gliding Chlamydomonas populations. Physical Rev. Res. 4:L042046 |
| Publication: | Ahmad R. et al. (2021) Light-Powered Reactivation of Flagella and Contraction of Microtubule Networks: Toward Building an Artificial Cell. ACS Synthetic Biology 10 (6):1490-1504 |
| Publication: | Ahmad R. et al. (2021) Light-Powered Reactivation of Flagella and Contraction of Microtubule Networks: Toward Building an Artificial Cell. ACS Synth Biol. 6 (10):1490-1504 |
| Publication: | Cammann J. et al. (2021) Emergent probability fluxes in confined microbial navigation. PNAS 118 (39):e2024752118 |
| Publication: | Clegg M.R. et al. (2021) Phenotypic Diversity and Plasticity of Photoresponse Across an Environmentally Contrasting Family of Phytoflagellates. Frontiers in Plant Science 12:707541 |
| Publication: | Fragkopoulos A.A. et al. (2021) Self-generated oxygen gradients control collective aggregation of photosynthetic microbes. J. R. Soc. Interface 18:20210553 |
| Publication: | Mojiri S. et al. (2021) Rapid multi-plane phase-contrast microscopy reveals torsional dynamics in flagellar motion. Biomed. Opt. Express 12 (6):3169-3180 |
| Publication: | Böddeker T.J. et al. (2020) Dynamic force measurements on swimming Chlamydomonas cells using micropipette force sensors. J. R. Soc. Interface 17 (162) |
| Publication: | Fragkopoulos A.A. et al. (2020) Light controls motility and phase separation of photosynthetic microbes. arXiv:2006.01675v1 |
| Publication: | Singh A.V. et al. (2020) Mechanical Coupling of Puller and Pusher Active Microswimmers Influences Motility. Langmuir 36 (19):5435-5443 |
| Publication: | Xu N. et al. (2020) Altered N-glycan composition impacts flagella mediated adhesion in Chlamydomonas reinhardtii. bioRxiv :2020.05.18.102624 |
| Publication: | Dunker S. (2019) Hidden Secrets Behind Dots: Improved Phytoplankton Taxonomic Resolution Using High-Throughput Imaging Flow Cytometry. Cytometry Part A 95 (8):854-868 |
| Publication: | Kreis C.T. et al. (2019) In vivo adhesion force measurements of Chlamydomonas on model substrates. Soft Matter 15 (14) |
| Publication: | Nowicka, B. (2019) Practical aspects of the measurements of non‐photochemical chlorophyll fluorescence quenching in green microalgae Chlamydomonas reinhardtii using Open FluorCam. Physiol. Plant. |
| Publication: | Bajerski F. et al. (2018) ATP Content and Cell Viability as Indicators for Cryostress Across the Diversity of Life. Front. Physiol. 9:921 |
| Publication: | Dunker S. et al. (2018) Combining high-throughput imaging, flow cytometry and deep learning for efficient species and life-cycle stage identification of phytoplankton. BMC Ecol. 18:51 |
| Publication: | Kreis C. et al. (2018) Adhesion of Chlamydomonas microalgae to surfaces is switchable by light. Nature Phys. 14:45-49 |
| Publication: | Ostapenko T. et al. (2018) Curvature-guided motility of microalgae in geometric confinement. Phys. Rev. Lett. 120:068002 |
| Publication: | Pröschold T. et al. (2018) Chlamydomonas schloesseri sp. nov. (Chlamydophyceae, Chlorophyta) revealed by morphology, autolysin cross experiments, and multiple gene analyses. Phytotaxa 362 (1):21-38 |
| Publication: | Saavedra R. et al. (2018) Comparative uptake study of arsenic, boron, copper, manganese and zinc from water by different green microalgae. Biores. Technol. 263:49-57 |
| Publication: | Santomauro G. et al. (2018) Incorporation of terbium into a microalga leads to magnetotactic swimmers. Advanced Biosystems 2 (12):1800039 |
| Publication: | Lachmann, Sabrina C. et al. (2016) Ecophysiology matters: linking inorganic carbon acquisition to ecological preference in four species of microalgae (Chlorophyceae). Journal of Phycology 52 (6):1051-1063 |
| Publication: | Krujatz F. et al. (2015) Green bioprinting: Viability and growth analysis of microalgae immobilized in 3D-plotted hydrogels versus suspension cultures. Eng. Life Sci. 15:678-688 |
| Publication: | Lode A. et al. (2015) Green bioprinting: Fabrication of photosynthetic algae-laden hydrogel scaffolds for biotechnological and medical applications. Eng. Life Sci. 15:177-183 |
| Publication: | Hallmann, A. (2011) Evolution of reproductive development in the volvocine algae. Sex Plant Reprod 24 (2):97-112 |
| Publication: | Lang I. et al. (2011) Fatty acid profiles and their distribution patterns in microalgae: a comprehensive analysis of more than 2000 strains from the SAG culture collection. BMC Plant Biol 11:124 |
| Publication: | Müller, J. et al. (2007) Assessing genetic stability of a range of terrestrial microalgae after cryopreservation using amplified fragment length polymorphism (AFLP). Am. J. Bot. 94 (5):799-808 |
| Publication: | Vieler A. et al. (2007) The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chemistry and Physics of Lipids 150:143–155 |
| Publication: | Pröschold, T. et al. (2005) Portrait of a species: Chlamydomonas reinhardtii. Genetics 170:1601-1610 |
| Publication: | Abe J. et al. (2004) The transcriptional program of synchronous gametogenesis in Chlamydomonas reinhardtii. Curr. Gen. 46:304-315 |
| Publication: | Buchheim, Mark A. et al. (1996) Phylogeny of the Chlamydomonadales (Chlorophyceae): A Comparison of Ribosomal RNA Gene Sequences from the Nucleus and the Chloroplast. Molecular Phylogenetics and Evolution 5 (2):391-402 |
| Publication: | Turmel M. et al. (1995) The trans-spliced intron 1 in the psaA gene of the Chlamydomonas chloroplast: a comparative analysis. Curr. Genet. 27:270-279 |
| Publication: | Turmel M. et al. (1993) Analysis of the Chloroplast Large Subunit Ribosomal RNA Gene from 17 Chlamydomonas Taxa Three Internal Transcribed Spacers and 12 Group I Intron Insertion Sites. Journal of Molecular Biology 232 (2):446-467 |
| Publication: | Kinoshita T. et al. (1992) Primary structure and expression of a gamete lytic enzyme in Chlamydomonas reinhardtii: similarity of functional domains to matrix metalloproteases. Proc. Natl. Acad. Sci. U.S.A. 89 (10):4693-4697 |
| Publication: | Kreimer G. et al. (1992) Functional analysis of the eyespot in Chlamydomonas reinhardtii mutant ey 627, mt (-). Planta 188 (4):513-21 |
| Publication: | Harris, E.H. (1989) The Chlamydomonas source book. A comprehensive guide to biology and laboratory use. |
| Publication: | Dolle R. et al. (1988) Effects of Calcium Ions and of Calcium Channel Blockers on Galvanotaxis of Chlamydomonas reinhardtii. Botanica Acta 101 (1):18-23 |
| Publication: | Klein U. et al. (1983) Photosynthetic Properties of Chloroplasts from Chlamydomonas reinhardii. Plant Physiol. 72 (2):488-491 |
| Publication: | Klein U. et al. (1983) Cellular Fractionation of Chlamydomonas reinhardii with Emphasis on the Isolation of the Chloroplast. Plant Physiol. 72 (2):481-487 |
| Publication: | Nultsch W. et al. (1975) Effect of external factors on phototaxis of Chlamydomonas reinhardtii. Archives of Microbiology 103 (1):175-179 |
| Publication: | Schlösser, U. (1967) Movie: Nuclear and Cell Division in the Volvocacea Chlamydomonas reinhardii. Institut für den Wissenschaftlichen Film |
| Publication: | Schlösser, U. (1966) Enzymatisch gesteuerte Freisetzung von Zoosporen bei Chlamydomonas reinhardtii Dangeard in Synchronkultur. Arch. Mikrobiol. 54:129-159 |
| Publication: | Hoshaw, R.W. (1965) Mating types of Chlamydomonas from the collection of Gilbert M. Smith. J. Phycol. 1 (4):194-196 |
| Publication: | Schlösser U. (1965) Enzymatisch gesteuerte Freisetzung von Zoosporen bei synchronisierter Chlamydomonas reinhardii Dangeard. |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequence Accession: | MG650074 Chlamydomonas reinhardtii SAG 11-32b ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. (1128 bp) |
| Sequence Accession: | MF678014 Chlamydomonas reinhardtii strain SAG 11-32b small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1 and 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2, partial sequence. (2390 bp) |
| Sequence Accession: | M94265 Chlamydomonas reinhardtii culture-collection SAG:11-32b cell wall lytic enzyme mRNA, complete cds. (2389 bp) |
| Sequence Accession: | AJ749629 5.8S rRNA gene, ITS1 and ITS2 (598 bp) |
| Sequence Accession: | D10542 Chlamydomonas reinhardtii mRNA for gamete lytic enzyme (GLE), complete cds. (2379 bp) |
| Sequence Accession: | U41442 Chlamydomonas reinhardtii ATP synthase subunit delta precursor (CREATPD) mRNA, nuclear gene encoding chloroplast protein, complete cds. (1286 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | yes |