| SAG Strain Number: | 1224-5/25 |
| TAXONOMY |
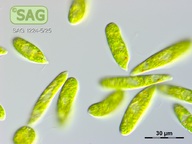
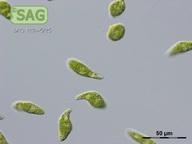
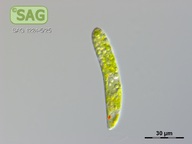
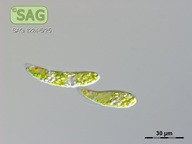
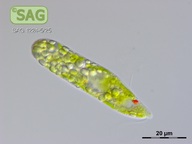
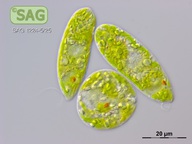
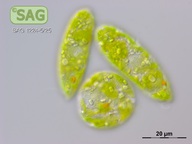
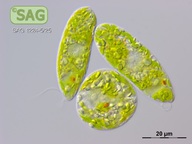
| Genus: | Euglena |
| Species: | gracilis |
| Taxonomic position: | Euglenophyta - Euglenophyceae |
| Authority: | Klebs |
| Variety: | |
| Formerly called: | |
| Authentic: | no |
| Division: | Euglenophyta |
| Class: | Euglenophyceae |
| ORIGIN |
| General habitat: | freshwater |
| Climatic zone: | |
| Continent: | |
| Country: | |
| Locality: | |
| Lat. / Long. (Precision): | |
| Year: | 1950 |
| Isolated by: | Pringsheim E.G. (leg. H. de Saedeleer, Belgium) |
| Strain number by isolator: | strain Z |
| Deposition by: | E.G. Pringsheim |
| Deposition date: | 1954 |
| CULTURE INFORMATION |
| Culture medium: | Eg Ag |
| Axenic: | axenity status under review. SAG 02/2022 |
| Strain relatives: | strain Z of E.G. Pringsheim; CCAP 1224/5Z; UTEX 753; ATCC 12894; UTCC 95 |
| Agitation resistance: | yes |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; used for bioassay of vitamin B12; recommended for schools |
| |
| PUBLICATIONS |
| Publication: | Gain G. et al. (2021) Trophic state alters the mechanism whereby energetic coupling between photosynthesis and respiration occurs in Euglena gracilis. New Phytol. 232:1603-1617 |
| Publication: | Krespach M.K.C. et al. (2020) Lichen-like association of Chlamydomonas reinhardtii and Aspergillus nidulans protects algal cells from bacteria. ISME J (14):2794-2805 |
| Publication: | Aiyar P. et al. (2017) Antagonistic bacteria disrupt calcium homeostasis and immobilize algal cells. Nat. Commun. 8 (1):1756 |
| Publication: | Dominik Cholewa (2017) Ein Bioraffineriekonzept mit Euglena gracilis. PhD Thesis. eDiss Link: https://pub.uni-bielefeld.de/record/2908500. |
| Publication: | Lakey, B. et al. (2017) The tetrapyrrole synthesis pathway as a model of horizontal gene transfer in euglenoids. J Phycol 53 (1):198–217 |
| Publication: | Grimm P. et al. (2015) Applicability of Euglena gracilis for biorefineries demonstrated by theproduction of alpha-tocopherol and paramylon followed by anaerobicdigestion. Journal of Biotechnology 215:72-79 |
| Publication: | Kim,J.I. et al. (2015) Taxon-rich multigene phylogeny of photosynthetic euglenoids (Euglenophyceae). Frontiers in Ecology and Evolution 3 |
| Publication: | Schwarzhans J.-P. et al. (2015) Dependency of the fatty acid composition ofEuglena gracilison growth phase and culture conditions. J. Appl. Phycol. 27:1389-1399 |
| Publication: | Milanowski R. et al. (2014) Distribution of Conventional and Nonconventional Introns in Tubulin (alpha and beta) Genes of Euglenids. Mol. Biol. Evol. 31 (3):584-593 |
| Publication: | Kim J.I. et al. (2010) Multigene Analyses of Photosynthetic Euglenoids and New Family, Phacaceae (Euglenales). J. Phycol. 46 (6):1278–1287 |
| Publication: | Tucci, S. et al. (2010) Variability of wax ester fermentation in natural and bleached Euglena gracilis Strains in response to oxygen and the elongase inhibitor flufenacet. J Eukaryot Microbiol 57 (1):63-9 |
| Publication: | Sara Tucci (2008) Wax ester fermentation and fatty acid biosynthesis in the facultatively anaerobic flagellate Euglena gracilis. PhD |
| Publication: | Schouten, Stefan et al. (2008) Evidence for substantial intramolecular heterogeneity in the stable carbon isotopic composition of phytol in photoautotrophic organisms. Organic Geochemistry 39 (1):135-146 |
| Publication: | Müller, J. et al. (2007) Assessing genetic stability of a range of terrestrial microalgae after cryopreservation using amplified fragment length polymorphism (AFLP). Am. J. Bot. 94 (5):799-808 |
| Publication: | Nowack E.C.M. et al. (2005) The 96-Well Twin-Layer System: A Novel Approach in the Cultivation of Microalgae. Protist 165 (2):239-251 |
| Publication: | Ryll, Joachim (2005) Fermentative Gewinnung von Paramylon aus Euglena gracilis auf Nebenprodukten der Stärkeindustrie in einer Pilotanlage. |
| Publication: | Felski, Michael (2004) Fermentative Gewinnung von Paramylon durch Euglena gracilis in konditioniertem Kartoffelfruchtwasser. |
| Publication: | Marin, B. et al. (2003) Phylogeny and taxonomic revision of plastid-containing Euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154 (1):99-145 |
| Publication: | Lebert M. et al. (1999) Negative gravitactic behavior of Euglena gracilis can not be described by the mechanism of buoyancy-oriented upward swimming. Advances in Space Research 24 (6):851-860 |
| Publication: | Henze K et al. (1995) A nuclear gene of eubacterial origin in Euglena gracilis reflects cryptic endosymbioses during protist evolution. PNAS 92 (20):9122-9126 |
| Publication: | Häder D.-P. et al. (1988) Inhibition of motility and phototaxis in the green flagellate, Euglena gracilis, by UV-B radiation. Arch. Microbiol. 150 (1):20-25 |
| Publication: | Häder D.-P. (1987) Polarotaxis, gravitaxis and vertical phototaxis in the green flagellate, Euglena gracilis. Arch. Microbiol. 147 (2):179-183 |
| Publication: | Cook J.R. (1982) ULTRAVIOLET INACTIVATION OF EUGLENA CHLOROPLASTS. III. EFFECT OF CULTURE AGE. Environmental and Experimental Botany 22 (2):175-179 |
| Publication: | Häder D.-P. et al. (1981) Phototaxis in the flagellates, Euglena gracilis and Ochromonas danica. Arch. Mikrobiol. 130 (1):78-82 |
| Publication: | Kivic, P.A. et al. (1974) Pinocytotic uptake of protein from the reservoir inEuglena. Arch. Microbiol. 96 (1):155-159 |
| Publication: | Koch W. (1973) Die algizide Wirkung chemischer Substanzen. Nachrichtenblatt Deutsch. Pflanzenschutzdienst 25:37-40 |
| Publication: | Pohl P. et al. (1972) Control of Fatty Acid and Lipid Biosynthesis in Euglena gracilis by Ammonia, Light and DCMU. Z. Naturforschung B 27 (b):53-61 |
| Publication: | Ohmann E. (1969) Die Regulation der Pyruvat-Kinase in Euglena gracilis . Arch. Mikrobiol. 67 (3):273-292 |
| Publication: | J.A. Gross et al.
(1960) Two c-Type Cytochromes from Light- and Dark-Grown Euglena. Science 132 (3423):357-358 |
| Publication: | Pringsheim E.G. et al. (1952) Experimental Elimination of Chromatophores and Eyespot in Euglena gracilis. New Phytologist 51 (1):65–76 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequencing Project: | PRJEB27422 The Euglena Genome Project |
| Sequence Accession: | MW646436 Euglena gracilis strain SAG1224-5/25-sq1 large subunit ribosomal RNA gene, partial sequence; chloroplast. (406 bp) |
| Sequence Accession: | L21904 Euglena gracilis culture-collection SAG:1224-5/25 glyceraldehyde-3-phosphate dehydrogenase (gapA) mRNA, complete cds. (1487 bp) |
| Sequence Accession: | L21903 Euglena gracilis culture-collection SAG:1224-5/25 glyceraldehyde-3-phosphate dehydrogenase (gapC) mRNA, complete cds. (1142 bp) |
| Sequence Accession: | L39772 Euglena gracilis culture-collection SAG:1224-5/25 glyceraldehyde-3-phosphate dehydrogenase (gapC) gene, complete cds. (4228 bp) |
| Sequence Accession: | AY741582 Euglena gracilis strain SAG1224-5/25 trans-2-enoyl-CoA reductase mRNA, complete cds. (1912 bp) |
| Sequence Accession: | KU500791 Euglena gracilis plastid uroporphyrinogen decarboxylase isoform 3 mRNA, partial cds; nuclear gene for plastid product. (720 bp) |
| Sequence Accession: | KU500760 Euglena gracilis plastid uroporphyrinogen decarboxylase isoform 1 mRNA, partial cds; nuclear gene for plastid product. (1344 bp) |
| Sequence Accession: | KU500718 Euglena gracilis plastid coproporphyrinogen oxidase isoform 4 mRNA, partial cds; nuclear gene for plastid product. (1179 bp) |
| Sequence Accession: | KU500717 Euglena gracilis plastid coproporphyrinogen oxidase isoform 3 mRNA, partial cds; nuclear gene for plastid product. (1380 bp) |
| Sequence Accession: | KU500692 Euglena gracilis plastid coproporphyrinogen oxidase isoform 2 mRNA, partial cds; nuclear gene for plastid product. (1155 bp) |
| Sequence Accession: | KU500691 Euglena gracilis plastid coproporphyrinogen oxidase isoform 1 mRNA, partial cds; nuclear gene for plastid product. (1137 bp) |
| Sequence Accession: | KU500666 Euglena gracilis plastid protoporphyrinogen oxidase mRNA, partial cds; nuclear gene for plastid product. (444 bp) |
| Sequence Accession: | KU500651 Euglena gracilis mitochondrial protoporphyrinogen oxidase mRNA, partial cds; nuclear gene for mitochondrial product. (444 bp) |
| Sequence Accession: | KU375875 Euglena gracilis plastid uroporphyrinogen synthase mRNA, partial cds; nuclear gene for plastid product. (450 bp) |
| Sequence Accession: | KU375859 Euglena gracilis plastid porphobilinogen synthase mRNA, partial cds; nuclear gene for plastid product. (1257 bp) |
| Sequence Accession: | KU375829 Euglena gracilis plastid porphobilinogen deaminase mRNA, partial cds; nuclear gene for plastid product. (1413 bp) |
| Sequence Accession: | KU375821 Euglena gracilis cytosolic porphobilinogen deaminase mRNA, partial cds. (363 bp) |
| Sequence Accession: | KU375800 Euglena gracilis plastid glutamyl tRNA reductase mRNA, partial cds; nuclear gene for plastid product. (1848 bp) |
| Sequence Accession: | KU375783 Euglena gracilis plastid glutamate 1-semialdehyde 2,1-aminotransferase mRNA, partial cds; nuclear gene for plastid product. (1656 bp) |
| Sequence Accession: | KU355529 Euglena gracilis cytosolic glutamyl tRNA synthetase mRNA, partial cds. (1635 bp) |
| Sequence Accession: | KU355501 Euglena gracilis plastid glutamyl tRNA synthetase mRNA, partial cds; nuclear gene for plastid product. (1839 bp) |
| Sequence Accession: | GQ925702 Euglena gracilis strain SAG 1224-5/25 chloroplast enolase precursor (eno29) gene, partial cds; nuclear gene for chloroplast product. (163 bp) |
| Sequence Accession: | GQ925703 Euglena gracilis strain SAG 1224-5/25 chloroplast ferredoxin precursor (petF) gene, partial cds; nuclear gene for chloroplast product. (413 bp) |
| Sequence Accession: | GQ925704 Euglena gracilis strain SAG 1224-5/25 chloroplast glyceraldehyde-3-phosphate dehydrogenase precursor (gapA) gene, partial cds; nuclear gene for chloroplast product. (320 bp) |
| Sequence Accession: | GQ925705 Euglena gracilis strain SAG 1224-5/25 chloroplast porphobilinogen deaminase precursor (pbgD) gene, partial cds; nuclear gene for chloroplast product. (430 bp) |
| Sequence Accession: | GQ925706 Euglena gracilis strain SAG 1224-5/25 chloroplast cytochrome f precursor (petA) gene, partial cds; nuclear gene for chloroplast product. (230 bp) |
| Sequence Accession: | GQ925707 Euglena gracilis strain SAG 1224-5/25 chloroplast cytochrome c6 precursor (petJ) gene, partial cds; nuclear gene for chloroplast product. (260 bp) |
| Sequence Accession: | GQ925708 Euglena gracilis strain SAG 1224-5/25 chloroplast photosystem I subunit F precursor (psaF) gene, partial cds; nuclear gene for chloroplast product. (350 bp) |
| Sequence Accession: | GQ925709 Euglena gracilis strain SAG 1224-5/25 chloroplast photosystem II protein M precursor (psbM) gene, partial cds; nuclear gene for chloroplast product. (420 bp) |
| Sequence Accession: | GQ925710 Euglena gracilis strain SAG 1224-5/25 chloroplast oxygen-evolving enhancer protein 1 precursor (psbO) gene, partial cds; nuclear gene for chloroplast product. (629 bp) |
| Sequence Accession: | GQ925711 Euglena gracilis strain SAG 1224-5/25 chloroplast photosystem II reaction center W precursor (psbW) gene, partial cds; nuclear gene for chloroplast product. (644 bp) |
| Sequence Accession: | KT591621 Euglena gracilis culture-collection SAG:1224525 plastid phosphoribulokinase mRNA, partial cds; nuclear gene for plastid product. (1098 bp) |
| Sequence Accession: | KT588873 Euglena gracilis culture-collection SAG:1224525 plastid glyceraldehyde 3-phosphate dehydrogenase mRNA, partial cds; nuclear gene for plastid product. (1380 bp) |
| Sequence Accession: | AJ272111 Euglena gracilis mRNA for enolase (eno02 gene). (1501 bp) |
| Sequence Accession: | AJ272112 Euglena gracilis partial mRNA for chloroplast enolase (eno29 gene). (1613 bp) |
| Sequence Accession: | AJ532426 Euglena gracilis partial 18S rRNA gene, strain SAG 1224-5/25. (2229 bp) |
| Sequence Accession: | AJ620503 Euglena gracilis mRNA for ribonucleotide reductase (rnr gene). (2458 bp) |
| Sequence Accession: | AJ620470 Euglena gracilis mRNA for dihydrolipoyl transacetylase. (1834 bp) |
| Sequence Accession: | AJ278425 Euglena gracilis mRNA for pyruvate:NADP+ oxidoreductase (pno gene). (5812 bp) |
| Sequence Accession: | AJ620471 Euglena gracilis mRNA for dihydrolipoyl dehydrogenase (lpd gene). (1811 bp) |
| Sequence Accession: | AJ620469 Euglena gracilis mRNA for pyruvate dehydrogenase E1 alpha subunit (pdh gene). (1586 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | yes |