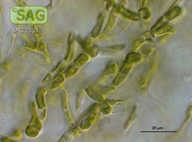
| SAG Strain Number: | 2039 |
| TAXONOMY |
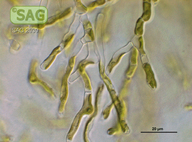
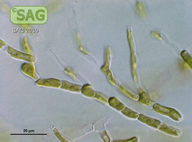
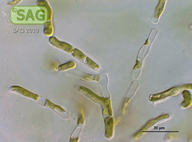
| Genus: | Ctenocladus |
| Species: | verrucariae |
| Taxonomic position: | Chlorophyta - Ulvophyceae |
| Authority: | Darienko et Pröschold 2022 |
| Variety: | |
| Formerly called: | non-authentic strain of Rindifilum verrucariae (Darienko et Pröschold) Malavasi et Škaloud 2022; non-authentic strain of Rindifilum ramosum gen. nov. et sp. nov. Malavasi, Klimešová, Lukešová et Škaloud 2022; Dilabifilum spec. |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Ulvophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN |
| General habitat: | phycobiont |
| Climatic zone: | temperate |
| Continent: | Europe |
| Country: | Germany |
| Locality: | Südschwarzwald, St. Wilhelmer Talbach, ca. 700 m, low water level, from lichen Verrucaria scabra |
| Lat. / Long. (Precision): | 47.885212 / 7.958808 (1500m) View on Google Maps |
| Year: | June 2000 |
| Isolated by: | H. Thüs |
| Strain number by isolator: | HT19 |
| Deposition by: | H. Thüs (< T. Friedl) |
| Deposition date: | before 03/2002 |
| CULTURE INFORMATION |
| Culture medium: | ES Ag |
| Axenic: | bacterial or other types of contamination |
| Strain relatives: | |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; strain SAG 2039 assigned as non-authentic strain of Ctenocladus verrucariae sp. nov. Darienko et Pröschold 2022, just after assigned as non-authentic strain of Rindifilum ramosum gen. nov. et spec. nov. Malavasi, Klimešová, Lukešová et Škaloud 2022; acc. to Malavasi et Skaloud 2022 strain has to be assigned as Rindifilum verrucariae (Darienko et Pröschold) Malavasi et Škaloud 2022. SAG 01/2024 |
| |
| PUBLICATIONS |
| Publication: | Darienko T. et al. (2022) The new photobiont Ctenocladus verrucariae sp. nov. (Ulvales, Ulvophyceae) found in various lichens of the family Verrucariaceae (Eurotiomycetes). Notulae Algarum 241:1-10 |
| Publication: | Malavasi V. et al. (2022) Rindifilum ramosum gen. nov., sp. nov., a new freshwater genus within the Ulvales (Ulvophyceae, Chlorophyta). Cryptogamie, Algologie 43 (8):125-133 |
| Publication: | del Campo E.M. et al. (2010) A single primer pair gives a specific ortholog amplicon in a wide range of Cyanobacteria and plastid-bearing organisms: applicability in inventory of reference material from collections and phylogenetic analysis. Mol. Phylogenet. Evol. 57 (3):1323-1328 |
| SEQUENCE INFORMATION |
| Sequence Accession: | OM994415 Ctenocladus verrucariae strain SAG 2039 elongation factor Tu (tufA) gene, partial cds; chloroplast. (855 bp) |
| Sequence Accession: | OM985641 Ctenocladus sp. TP-2022a strain SAG 2039 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and large subunit ribosomal RNA gene, partial s (2427 bp) |
| Sequence Accession: | MF000574 Pseudopleurococcus sp. A SAG 2039 18S ribosomal RNA gene, partial sequence. (1713 bp) |
| Sequence Accession: | MF000593 Pseudopleurococcus sp. A SAG 2039 elongation factor Tu (tufA) gene, partial cds; chloroplast. (804 bp) |
| Sequence Accession: | HQ003299 Pseudopleurococcus sp. A SAG 2039 culture SAG:2039 large subunit ribosomal RNA gene, partial sequence; chloroplast. (773 bp) |
| Sequence Accession: | JQ921006 Pseudopleurococcus sp. A SAG 2039 23S large subunit ribosomal RNA gene, partial sequence; exon; putative LAGLIDADG homing endonuclease gene, complete cds; exon; putative LAGLIDADG homing endonuclease gene, complete cds; exons; putative LAGLIDADG homi (5356 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | no |