| SAG Strain Number: | 211-11b |
| TAXONOMY |
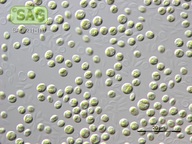
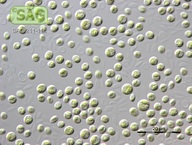
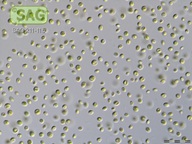
| Genus: | Chlorella |
| Species: | vulgaris |
| Taxonomic position: | Chlorophyta - Trebouxiophyceae |
| Authority: | Beijerinck |
| Variety: | |
| Formerly called: | |
| Authentic: | yes |
| Division: | Chlorophyta |
| Class: | Trebouxiophyceae |
| ORIGIN |
| General habitat: | freshwater |
| Climatic zone: | temperate |
| Continent: | Europe |
| Country: | Netherlands |
| Locality: | eutrophic shallow pond near Delft |
| Lat. / Long. (Precision): | 52.011513 / 4.358740 (20000m) View on Google Maps |
| Year: | April 1889 |
| Isolated by: | M.W. Beijerinck |
| Strain number by isolator: | as Chlorella vulgaris, later Delft No. 35 as C. miniata |
| Deposition by: | E.G. Pringsheim (< Meyer Prague Nr. 35 < Westerdijk Delft) |
| Deposition date: | 1954 |
| CULTURE INFORMATION |
| Culture medium: | ESP Ag |
| Axenic: | axenic |
| Strain relatives: | CCAP 211/11B; UTEX 259 |
| Agitation resistance: | yes |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; potential for Daphnia sp. feeding; oldest strain of the SAG; potential test strain for ecotoxicity testing (DIN 38412, Teil 33 (DEV L 33); ISO 8692 (1989); OECD 201 (2011)) |
| |
| PUBLICATIONS |
| Publication: | Gao Y. et al. (2024) The effect of light intensity on microalgae biofilm structures and physiology under continuous illumination. Sci. Rep. 14:1151 |
| Publication: | Pozzobon V. et al. (2024) Microalgal cell division tracking using CFSE. Algal Res. 80:103501 |
| Publication: | Pozzobon V. et al. (2024) Systematic formulation of brewery effluent for high-efficiency epuration and food-grade microalgae production. 10.1016/j.biteb.2024.101795 25:101795 |
| Publication: | Pozzobon V. et al. (2024) Chlorella vulgaris cold preservation (4degrC) as a means to stabilize biomass for bioreactor inoculation: A six-month study. Aquacult. Engin. 107:102449 |
| Publication: | Rautenberger R. et al. (2024) Growth strategies of Chlorella vulgaris in seawater for a high production of biomass and lipids suitable for biodiesel. Algal Res. 77:103360 |
| Publication: | Weithoff G. et al. (2024) Weak effect of temperature fluctuations on the invasion of Raphidiopsis raciborskii (Cyanobacteria) in experimental plankton microcosms. J. Phycol. 61:261-266 |
| Publication: | Brückner K. et al. (2023) Permeabilization of the cell wall of Chlorella sorokiniana by the chitosan-degrading protease papain. Algal Res. 71:103066 |
| Publication: | Gao Y. et al. (2023) The impact of light/dark regimes on structure and physiology of Chlorella vulgaris biofilms. Front. Microbiol., Sec. Aquatic Microbiology 14:1250866 |
| Publication: | Kryvenda A. et al. (2023) Testing for terrestrial and freshwater microalgae productivity under elevated CO2 conditions and nutrient limitation. BMC Plant. Biol. 23:27 |
| Publication: | Levasseur W. et al. (2023) Chlorella vulgaris acclimated cultivation under flashing light: An in-depth investigation under iso-actinic conditions. Algal Res. 70:102976 |
| Publication: | Oliver A. et al. (2023) Assessment of Photosynthetic Carbon Capture versus Carbon Footprint of an Industrial Microalgal Process. Appl. Sci. 13:5193 |
| Publication: | Pozzobon V. et al. (2023) Impact of 3D printing materials on microalga Chlorella vulgaris. Bioresource Technol. 389:129807 |
| Publication: | Schreiber U. (2023) Light-induced changes of far-red excited chlorophyll fluorescence: Further evidence for variable chlorophyll fluorescence of photosystem I in vivo. Photosynth. Res. |
| Publication: | Pozzobon V. (2022) Chlorella vulgaris cultivation under super high light intensity: An application of the flashing light effect. Algal Res. 68:102874 |
| Publication: | Liyanaarachchi V.C. et al. (2021) Artificial neural network (ANN) approach to optimize cultivation conditions of microalga Chlorella vulgaris in view of biodiesel production. Biochemical Engineering Journal 173:108072 |
| Publication: | Pozzobon V. et al. (2021) Nitrate and nitrite as mixed source of nitrogen for Chlorella vulgaris: Growth, nitrogen uptake and pigment contents. Bioresource Technol. 330:124995 |
| Publication: | Schreiber U. et al. (2021) Evidence for variable chlorophyll fluorescence of photosystem I in vivo. Photosynth. Res. 149:213-231 |
| Publication: | Aigner S. et al. (2020) Adaptation to Aquatic and Terrestrial Environments in Chlorella vulgaris (Chlorophyta). Front. Microbiol. 11:2451 |
| Publication: | Lewington-Pearce L. et al. (2020) Diversity and temperature indirectly reduce CO2 concentrations in experimental freshwater communities. Oecologia 192:515-527 |
| Publication: | Mócsai R. et al. (2020) A first view on the unsuspected intragenus diversity of N‐glycans in Chlorella microalgae. Plant J. 103:184-196 |
| Publication: | Pozzobon V. et al. (2020) Machine learning processing of microalgae flow cytometry readings: illustrated with Chlorella vulgaris viability assays. J. Appl. Phycol. |
| Publication: | Pulgarin A. et al. (2020) A novel proposition for a citrate-modified photo-Fenton process against bacterial contamination of microalgae cultures. Applied Catalysis B: Environmental 265:118615 |
| Publication: | Widzgowski J. et al. (2020) High light induces species specific changes in the membrane lipid composition of Chlorella. Biochem. J. 477 (13):2543-2559 |
| Publication: | Darienko T. et al. (2019) Are there any true marine Chlorella species? Molecular phylogenetic assessment and ecology of marine Chlorella-like organisms, including a description of Droopiella gen. nov. Systematics and Biodiversity 17 (8):811-829 |
| Publication: | Dunker S. (2019) Hidden Secrets Behind Dots: Improved Phytoplankton Taxonomic Resolution Using High-Throughput Imaging Flow Cytometry. Cytometry Part A 95 (8):854-868 |
| Publication: | Mócsai R. et al. (2019) N-glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Sci. Rep. 9:331 |
| Publication: | Sakarika M. et al. (2019) Chlorella vulgaris as a green biofuel factory: Comparison between biodiesel, biogas and combustible biomass production. Bioresource Technol. 273:237-243 |
| Publication: | Schreiber U. et al. (2019) Rapidly reversible chlorophyll fluorescence quenching induced by pulses of supersaturating light in vivo. Photosynth. Res. 142:35-50 |
| Publication: | Bajerski F. et al. (2018) ATP Content and Cell Viability as Indicators for Cryostress Across the Diversity of Life. Front. Physiol. 9:921 |
| Publication: | Dunker S. et al. (2018) Combining high-throughput imaging, flow cytometry and deep learning for efficient species and life-cycle stage identification of phytoplankton. BMC Ecol. 18:51 |
| Publication: | Dunker S. et al. (2018) Cell Wall Structure of Coccoid Green Algae as an Important Trade-Off Between Biotic Interference Mechanisms and Multidimensional Cell Growth. Frontiers in Microbiology 9:719 |
| Publication: | Sakarika M. et al. (2017) Kinetics of growth and lipids accumulation in Chlorella vulgaris during batch heterotrophic cultivation: Effect of different nutrient limitation strategies. Bioresour Technol 243:356-365 |
| Publication: | Gruber-Brunhumer M.R. et al. (2016) Associated effects of storage and mechanical pre-treatments of microalgae biomass on biomethane yields in anaerobic digestion. Biomass and Bioenergy 93:259-268 |
| Publication: | Hodac, L. et al. (2016) Widespread green algae Chlorella and Stichococcus exhibit polar-temperate and tropical-temperate biogeography. FEMS Microbiol Ecol 92 (8) |
| Publication: | Markou, Giorgos et al. (2016) Applying raw poultry litter leachate for the cultivation of Arthrospira platensis and Chlorella vulgaris. Algal Research 13:79-84 |
| Publication: | Morschett, Holger et al. (2016) Simplified cryopreservation of the microalga Chlorella vulgaris integrating a novel concept for cell viability estimation. Engineering in Life Sciences 16 (1):36-44 |
| Publication: | Postma P.R. et al. (2016) Selective extraction of intracellular components from the microalga Chlorella vulgaris by combined pulsed electric field–temperature treatment. Bioresource Technol. 203:80-88 |
| Publication: | Sakarika M. et al. (2016) Effect of pH on growth and lipid accumulation kinetics of the microalga Chlorella vulgaris grown heterotrophically under sulfur limitation. Bioresource Technol. 219:694-701 |
| Publication: | Steudel B. et al. (2016) Contrasting biodiversity-ecosystem functioning relationships in phylogenetic and functional diversity. New Phytologist 212 (2 ): 409-420 |
| Publication: | Straessner R. et al. (2016) icroalgae precipitation in treatment chambers during pulsed electric field (PEF) processing. Innov. Food Sci. Emerg. Technol. 37:391-399 |
| Publication: | Torri C. et al. (2016) Fast Procedure for the Analysis of Hydrothermal Liquefaction Biocrude with Stepwise Py-GC-MS and Data Interpretation Assisted by Means of Non-negative Matrix Factorization. Energy & Fuels 30 (2):1135-1144 |
| Publication: | Vandamme D. et al. (2016) Inhibition of alkaline flocculation by algal organic matter for Chlorella vulgaris. Water Res 88:301-7 |
| Publication: | Beuckels A. et al. (2015) Nitrogen availability influences phosphorus removal in microalgae-based wastewater treatment. Water Research 77:98-106 |
| Publication: | Heeg J.S. et al. (2015) ITS2 and 18S rDNA sequence-structure phylogeny of Chlorella and allies (Chlorophyta, Trebouxiophyceae, Chlorellaceae). Plant Gene 4:20-28 |
| Publication: | Klughammer C. et al. (2015) Apparent PS II absorption cross-secion and estimation of mean PAR in optically thin and dense suspensions of Chlorella. Photosynth. Res. 123 (1):77-92 |
| Publication: | Krienitz, L. et al. (2015) Chlorella: 125 years of the green survivalist. Trends Plant Sci 20 (2):67-9 |
| Publication: | Rajevic, N. et al. (2015) Algal endosymbionts in European Hydra strains reflect multiple origins of the zoochlorella symbiosis. Molecular Phylogenetics and Evolution 93:55-62 |
| Publication: | Rajevic, N. et al. (2015) Algal endosymbionts in European Hydra strains reflect multiple origins of the zoochlorella symbiosis. Molecular Phylogenetics and Evolution 93:55-62 |
| Publication: | Goiris K. et al. (2014) Detection of flavonoids in microalgae from different evolutionary lineages. J. Phycol. 50 (3):483-492 |
| Publication: | Mudimu O. et al. (2014) Biotechnological screening of microalgal and cyanobacterial strains for biogas production and antibacterial and antifungal effects. Metabolites 4 (2):373-393 |
| Publication: | Prakash Binnal et al. (2014) Lipid productivity of microalgae Chlorella vulgaris and Nannochloropsis oculata in externally illuminated lab scale photobioreactor. International Journal of ChemTech Research 7 (5):2217-2221 |
| Publication: | Salim S. et al. (2014) Mechanism behind autoflocculation of unicellular green microalgae Ettlia texensis. Journal of Biotechnology 174:34-38 |
| Publication: | López Barreiro D. et al. (2013) Influence of strain-specific parameters on hydrothermal liquefaction of microalgae. Bioresource Technology 146:463-471 |
| Publication: | Schreiber U. et al. (2013) Wavelength-dependent photodamage to Chlorella investigated with a new type of multi-color PAM chlorophyll fluorometer. Photosynth. Res. 114:165-177 |
| Publication: | Schreiber U. et al. (2012) Assessment of wavelength-dependent parameters of photosynthetic electron transport with a new type of multi-color PAM chlorophyll fluorometer. Photosynth. Res. 113:127-144 |
| Publication: | Steudel B. et al. (2012) Biodiversity effects on ecosystem functioning change along environmental stress gradients. Ecology Letters 15 (12):1397–1405 |
| Publication: | Blache U. et al. (2011) The impact of cell-specific absorption properties on the correlation of electron transport rates measured by chlorophyll fluorescence and photosynthetic oxygen production in planktonic algae. Plant Physiol. Biochem. 49 (8):801-808 |
| Publication: | Bock C., Krienitz L. et al. (2011) Taxonomic reassessment of the genus Chlorella (Trebouxiophyceae) using molecular signatures (barcodes), including description of seven new species. Fottea 11 (2):293–312 |
| Publication: | Gustavs et al. (2011) POLYOL PATTERNS IN BIOFILM-FORMING AEROTERRESTRIAL GREEN ALGAE (TREBOUXIOPHYCEAE, CHLOROPHYTA). J. Phycol. 47:533-537 |
| Publication: | Ryckebosch E. et al. (2011) Optimization of an Analytical Procedure for Extraction of Lipids from Microalgae. Journal of the American Oil Chemists' Society 89 (2):189-198 |
| Publication: | Salim S. et al. (2011) Harvesting of microalgae by bio-flocculation. J Appl Phycol 23 (5):849-855 |
| Publication: | Vandamme D. et al. (2011) Evaluation of electro-coagulation-flocculation for harvesting marine and freshwater microalgae. Biotechnol Bioeng 108 (10):2320-9 |
| Publication: | Vogel, Manja (2011) Zur Aufnahme und Bindung von Uran(VI) durch die Grünalge Chlorella vulgaris. |
| Publication: | Görs M. et al. (2010) THE POTENTIAL OF ERGOSTEROL AS CHEMOTAXONOMIC MARKER TO DIFFERENTIATE BETWEEN ‘‘CHLORELLA’’ SPECIES (CHLOROPHYTA). J. Phycol. 46:1296–1300 |
| Publication: | Lapaille et al. (2010) Atypical Subunit Composition of the Chlorophycean Mitochondrial F1FO-ATP Synthase and Role of Asa7 Protein in Stability and Oligomycin Resistance of the Enzyme. 27 (7):1630-1644 |
| Publication: | Pröschold, T. et al. (2010) Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycol. Res. 58 (1):1-8 |
| Publication: | Vogel M. et al. (2010) Biosorption of U(VI) by the green algae Chlorella vulgaris in dependence of pH value and cell activity. Science of The Total Environment 409 (2):384–395 |
| Publication: | Görs M. et al. (2009) Quality analysis of commercial Chlorella products used as dietary supplement in human nutrition. Journal of Applied Phycology 22 (3):265-276 |
| Publication: | Kamjunke, Norbert et al. (2008) ALGAE AS COMPETITORS FOR GLUCOSE WITH HETEROTROPHIC BACTERIA. Journal of Phycology 44 (3):616-623 |
| Publication: | Vogel M. et al. (2008) Microscopic and spectroscopic investigation of U(VI) interaction with monocellular green algae. In: Merkel, B.J. et al. [Eds.] Uranium, Mining and Hydrogeology |
| Publication: | John G. Day J.G. et al. (2007) The Use of Physical and Virtual Infrastructures for the Validation of Algal Cryopreservation Methods in International Culture Collections. CryoLetters 28 (5):359-376 |
| Publication: | Müller, J. et al. (2007) Assessing genetic stability of a range of terrestrial microalgae after cryopreservation using amplified fragment length polymorphism (AFLP). Am. J. Bot. 94 (5):799-808 |
| Publication: | Luo W. et al. (2006) Genotype versus Phenotype Variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 157 (3):315-333 |
| Publication: | Shi J. et al. (2006) Removal of nitrogen and phosphorus from wastewater using microalgae immobilized on twin layers: an experimental study. J. Appl. Phycol. 19 (5):417-423 |
| Publication: | Habetha, M. et al. (2005) Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. J Exp Biol 208 (Pt 11):2157-65 |
| Publication: | Müller J. et al. (2005) Distinction between multiple isolates of Chlorella vulgaris (Chlorophyta, Trebouxiophyceae) and testing for conspecificity using amplified fragment length polymorphism and its rDNA sequences. J. Phycol. 41 (6):1236-1247 |
| Publication: | Krienitz,L. et al. (2004) Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae). Phycologia 43 (5):529-542 |
| Publication: | Friedl, T. et al. (2002) Phylogenetic relationships of green algae assigned to the genus Planophila (Chlorophyta): evidence from 18S rDNA sequence data and ultrastructure. Eur. J. Phycol. 37:373-384 |
| Publication: | Nemcova Y. et al. (2000) Cell wall development, microfibril and pyrenoid structure in type strains of Chlorella vulgaris, C. kessleri, C. sorokiniana compared with C. luteoviridis (Trebouxiophyceae, Chlorophyta). Arch. Hydrobiol. Suppl. (Algol. Stud. 100) 136:95-105 |
| Publication: | Linz, B. et al. (1999) Correlation between virus-sensitivity and isoenzyme spectrum in symbiotic Chlorella-like algae. Protistology 1 (2):76-81 |
| Publication: | Huss V.A.R. et al. (1993) There is an ecological basis for host/symbiont specificity in Chlorella/Hydra symbioses. Endocyt. Cell Res. 10 (1-2):35-46 |
| Publication: | Huss, V.A. et al. (1990) Phylogenetic position of some Chlorella species within the chlorococcales based upon complete small-subunit ribosomal RNA sequences. J. Mol. Evol. 31 (5):432-442 |
| Publication: | Huss V.A. et al. (1989) Primary structure of the chloroplast small subunit ribosomal RNA gene from Chlorella vulgaris. Nucleic Acids Res. 17 (22):9487 |
| Publication: | Huss V.A. et al. (1989) Primary structure of the Chlorella vulgaris small subunit ribosomal RNA coding region. Nucleic Acids Res. 17 (3):1255 |
| Publication: | Kessler E. (1986) Limits of growth of five Chlorella species in the presence of toxic heavy metals. Arch. Hydrobiol. Suppl. (Algological Studies 42) 73:123-128 |
| Publication: | Rahat M. et al. (1986) Algal endosymbiosis in brown hydra: host/symbiont specificity. J. Cell Sci. 86:273-286 |
| Publication: | Reisser W. et al. (1986) Studies on ultrastructure and host range of a Chlorella attacking virus. Protoplasma 135 (2):162-165 |
| Publication: | Piorreck M. et al. (1984) Biomass production, total protein, chlorophylls, lipids and fatty acids of freshwater green and blue-green algae under different nitrogen regimes. Phytochemistry 23 (2):207-216 |
| Publication: | Reisser, Werner (1984) The taxonomy of green algae endosymbiotic in ciliates and a sponge. 19:309-318 |
| Publication: | Niess, Dörte et al. (1982) Photobehaviour of Paramecium bursaria infected with different symbiotic and aposymbiotic species of Chlorella. Planta 156 (5):475-480 |
| Publication: | Kessler E. (1976) Comparative physiology, biochemistry, and the taxonomy of Chlorella (Chlorophyceae). Plant Systematics and Evolution 125 (3):129-138 |
| Publication: | Liersch R. (1976) Über das ätherische Öl von Grünalgen. I. Die Öle der Gattung Chlorella (On the essential oil of green algae. I. The oils of the genus Chlorella). Arch. Microbiol. 107:353-356 |
| Publication: | Pohl P. et al. (1975) Stickstoff-Ausscheidung durch N 2-fixierende Blaualgen, III Verwertung des ausgeschiedenen Stickstoffs durch Grünalgen (Nitrogen Excretion by Nitrogen Fixing Blue-green Algae, III Utilization of the Excreted Nitrogen by Green Algae). Z. Naturforschung 30 (c):223-226 |
| Publication: | Pohl P. et al. (1971) Über den Einfluss von anorganischem Stickstoff-Gehalt in der Nährlösung auf die Fettsäure-Biosynthese in Grünalgen. Phytochemistry 10 (7):1505-1513 |
| Publication: | Kessler E. (1967) Physiologische und biochemische Beiträge zur Taxonomie der Gattung Chlorella III. Merkmale von 8 autotrophen Arten. Archiv für Mikrobiologie 55 (4):346-357 |
| Publication: | Kessler, E. et al. (1962) Biochemical Contributions to the Taxonomy of the Genus Chlorella. Nature 194:1096-1097 |
| Publication: | Pringsheim E.G. (1951) Die Sammlung von Algen-, Flagellaten- und Mooskulturen am Botanischen Institut der Universität Cambridge. Culture collection of algae and protozoa, botany school, University of Cambridge, England. Archiv. Mikrobiol. 16:1-17 |
| Publication: | Beijerinck M.W. (1890) Culturversuche mit Zoochlorellen, Lichenengonidien und anderen niederen Algen. Z. Bot. 45-4:725-785 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequence Accession: | AY323465 Chlorella vulgaris strain SAG 211-11b internal transcribed spacer 2, complete sequence. (259 bp) |
| Sequence Accession: | AF499684 Chlorella vulgaris strain SAG 211-11b voucher SAG:211-11b ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. (1358 bp) |
| Sequence Accession: | AY591508 Chlorella vulgaris strain SAG 211-11b 18S ribosomal RNA gene, partial sequence; internal transcribed spacer 1, 5.8S ribosomal RNA gene, and internal transcribed spacer 2, complete sequence; and 26S ribosomal RNA gene, partial sequence. (765 bp) |
| Sequence Accession: | X13688 18S ribosomal RNA gene (1798 bp) |
| Sequence Accession: | FM205832 Chlorella vulgaris 18S rRNA gene (partial), ITS1, 5.8S rRNA gene, ITS2 and 28S rRNA gene (partial), strain SAG 211-11b. (2496 bp) |
| Sequence Accession: | FR694223 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MB. (1368 bp) |
| Sequence Accession: | FR694222 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MAC. (1368 bp) |
| Sequence Accession: | FR694221 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MA21. (1368 bp) |
| Sequence Accession: | FR694220 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MA24. (1357 bp) |
| Sequence Accession: | FR694219 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MB16. (976 bp) |
| Sequence Accession: | FR694218 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MB13. (952 bp) |
| Sequence Accession: | FR694217 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MAA. (741 bp) |
| Sequence Accession: | FR694216 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MA20. (992 bp) |
| Sequence Accession: | FR694215 Chlorella vulgaris 5S rRNA gene (partial), IGS and 26S rRNA gene (partial), specimen voucher SAG:211-11b, clone Ac15I1MB12. (1020 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | good survival |
| TEACHING |
| Recommended for teaching: | yes |