| SAG Strain Number: | 34-1b |
| TAXONOMY |
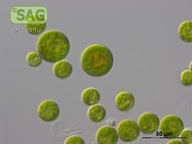
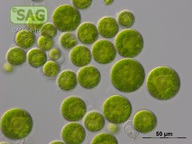
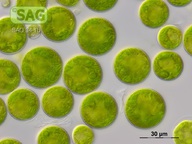
| Genus: | Haematococcus |
| Species: | pluvialis |
| Taxonomic position: | Chlorophyta - Chlorophyceae |
| Authority: | Flotow em. Wille |
| Variety: | |
| Formerly called: | H. lacustris |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN |
| General habitat: | freshwater |
| Climatic zone: | |
| Continent: | Europe |
| Country: | former Czechoslovakia |
| Locality: | |
| Lat. / Long. (Precision): | |
| Year: | probably 1931 |
| Isolated by: | F. Mainx |
| Strain number by isolator: | |
| Deposition by: | E.G. Pringsheim ( < F. Mainx before 1939) |
| Deposition date: | 1954 |
| CULTURE INFORMATION |
| Culture medium: | ESP Ag |
| Axenic: | axenic |
| Strain relatives: | UTEX 16; ATCC 30402 |
| Agitation resistance: | yes |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; recommended for schools |
| |
| PUBLICATIONS |
| Publication: | Kryvenda A. et al. (2023) Testing for terrestrial and freshwater microalgae productivity under elevated CO2 conditions and nutrient limitation. BMC Plant. Biol. 23:27 |
| Publication: | Enrico Ehrhardt (2022) Physiologische und molekulare Vorgänge während des Übergangs von Haematococcus pluvialis in das nicht-mobile, Astaxanthin-bildende Zellstadium. |
| Publication: | Roach T. et al. (2022) Acquisition of desiccation tolerance in Haematococcus pluvialis requires photosynthesis and coincides with lipid and astaxanthin accumulation. Algal Res. 64:102699 |
| Publication: | Kraus D. et al. (2021) Three step flow focusing enables image-based discrimination and sorting of late stage 1 Haematococcus pluvialis cells. PLOS ONE |
| Publication: | Sascha Baer (2018) Screening und Optimierung von Produktionsorganismen für mikroalgenbasierte Bioraffinerieverfahren. |
| Publication: | Alanagreh L. et al. (2017) Assessing intragenomic variation of the internal transcribed spacer two: Adapting the Illumina metagenomics protocol. PLoSOne 12 (7):e0181491 |
| Publication: | Kiperstok, A. C. et al. (2017) Biofilm cultivation of Haematococcus pluvialis enables a highly productive one-phase process for astaxanthin production using high light intensities. Algal Research-Biomass Biofuels and Bioproducts 21:213-222 |
| Publication: | Nakada T. et al. (2016) What is the correct name for the type of Haematococcus Flot. (Volvocales, Chlorophyceae)?. Taxon 65:343-348 |
| Publication: | Taucher J. et al. (2016) Cell Disruption and Pressurized Liquid Extraction of Carotenoids from Microalgae. J. Thermodyn. Catal. 7 (1):1000158 |
| Publication: | Céline C. Allewaert et al. (2015) Species diversity in European Haematococcus pluvialis (Chlorophyceae, Volvocales). Phycologia 54 (6):583-598 |
| Publication: | Lemieux C. et al. (2015) Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol 15:264 |
| Publication: | Buchheim, M.A. et al. (2013) The blood alga: phylogeny of Haematococcus (Chlorophyceae) inferred from ribosomal RNA gene sequence data. Eur. J. Phycol. 48 (3):318-329 |
| Publication: | Han, Danxiang et al. (2012) SUSCEPTIBILITY AND PROTECTIVE MECHANISMS OF MOTILE AND NON MOTILE CELLS OF HAEMATOCOCCUS PLUVIALIS (CHLOROPHYCEAE) TO PHOTOOXIDATIVE STRESS1. Journal of Phycology 48 (3):693-705 |
| Publication: | Collins, AM et al. (2011) Carotenoid Distribution in Living Cells of Haematococcus pluvialis (Chlorophyceae). PLoS ONE 6 (9):e24302 |
| Publication: | Friedl, T. et al. (2002) Phylogenetic relationships of green algae assigned to the genus Planophila (Chlorophyta): evidence from 18S rDNA sequence data and ultrastructure. Eur. J. Phycol. 37:373-384 |
| Publication: | Hepperle, D. et al. (1998) Phylogenetic position of the Phacotaceae within the Chlamydophyceae as revealed by analysis of 18S rDNA- and rbcL- sequences. J. Mol. Evol. 47 (4):420-430 |
| Publication: | Buchheim, Mark A. et al. (1996) Phylogeny of the Chlamydomonadales (Chlorophyceae): A Comparison of Ribosomal RNA Gene Sequences from the Nucleus and the Chloroplast. Molecular Phylogenetics and Evolution 5 (2):391-402 |
| Publication: | Pringsheim, E.G. (1966) Nutritional requirements of Haematococcus pluvialis and related species. J. Phycol. 2 (1):1-7 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequencing Project: | PRJNA827483 Comparative De novo Transcriptomics and Random UV mutagenesis: Application in Astaxanthin Production Enhancement for Haematococcus pluvialis |
| Sequence Accession: | OL711901 Haematococcus lacustris isolate SAG 34-1b large subunit ribosomal RNA gene, partial sequence. (2036 bp) |
| Sequence Accession: | OL699992 Haematococcus lacustris isolate SAG 34-1b internal transcribed spacer 1, partial sequence. (326 bp) |
| Sequence Accession: | OL739477 Haematococcus lacustris isolate SAG 34-1b 5.8S ribosomal RNA gene, partial sequence. (155 bp) |
| Sequence Accession: | MF163210 Haematococcus lacustris isolate SAG34_1b_H6 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163209 Haematococcus lacustris isolate SAG34_1b_H5 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163208 Haematococcus lacustris isolate SAG34_1b_H4 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163207 Haematococcus lacustris isolate SAG34_1b_H3 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163206 Haematococcus lacustris isolate SAG34_1b_H2 internal transcribed spacer 2, partial sequence. (226 bp) |
| Sequence Accession: | MF163205 Haematococcus lacustris isolate SAG34_1b_H1 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163204 Haematococcus lacustris isolate SAG34_1b clone CS004_11 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163203 Haematococcus lacustris isolate SAG34_1b clone CS004_09 internal transcribed spacer 2, partial sequence. (226 bp) |
| Sequence Accession: | MF163202 Haematococcus lacustris isolate SAG34_1b clone CS004_05 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | MF163201 Haematococcus lacustris isolate SAG34_1b clone CS004_02 internal transcribed spacer 2, partial sequence. (226 bp) |
| Sequence Accession: | MF163200 Haematococcus lacustris isolate SAG34_1b clone CS004_01 internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | AF159369 Haematococcus lacustris strain SAG 34-1b culture-collection SAG:34-1b 18S ribosomal RNA gene, complete sequence. (1748 bp) |
| Sequence Accession: | KC153465 Haematococcus lacustris isolate SAG34_1b internal transcribed spacer 2, partial sequence. (227 bp) |
| Sequence Accession: | L49151 Haematococcus lacustris chloroplast large-subunit ribosomal RNA (rrnL) gene. (6713 bp) |
| Sequence Accession: | KT625205 Haematococcus lacustris culture-collection SAG:34-1b clone contig-1 chloroplast, partial genome. (8490 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | good survival, standard 2-step protocol 5% DMSOor 5% MetOH |
| TEACHING |
| Recommended for teaching: | yes |