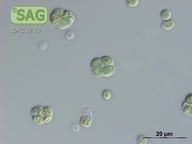
| SAG Strain Number: | 39.79 |
| TAXONOMY |
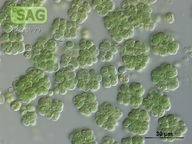
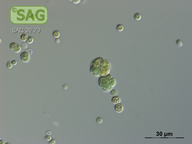
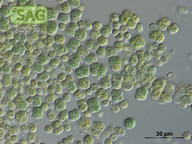
| Genus: | Chroococcidiopsis |
| Species: | cubana |
| Taxonomic position: | Cyanobacteria |
| Authority: | Komárek et Hindák |
| Variety: | |
| Formerly called: | C. thermalis |
| Authentic: | no |
| Division: | Cyanobacteria |
| Class: | Cyanophyceae |
| ORIGIN |
| General habitat: | soil |
| Climatic zone: | tropic |
| Continent: | America |
| Country: | Cuba |
| Locality: | Pinar del Rio, dry soil |
| Lat. / Long. (Precision): | 22.423248 / -83.69113 (8000m) View on Google Maps |
| Year: | 1966 |
| Isolated by: | F. Hindák |
| Strain number by isolator: | 1966/27 |
| Deposition by: | F. Hindák |
| Deposition date: | before 1979 |
| CULTURE INFORMATION |
| Culture medium: | BG11 Ag |
| Axenic: | axenic |
| Strain relatives: | ATCC 29381; PCC 7433; DSM 107010 |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain |
| |
| PUBLICATIONS |
| Publication: | Jung P.J. et al. (2024) The underestimated fraction: diversity, challenges and novel insights into unicellular cyanobionts of lichens. ISME Comm. 4 (1):ycae069 |
| Publication: | Antonaru L.A. et al. (2023) Common loss of far-red light photoacclimation in cyanobacteria from hot and cold deserts: a case study in the Chroococcidiopsidales. ISME Comm. 3 (1):113 |
| Publication: | Will, S. et al. (2018) Day and Night: Metabolic Profiles and Evolutionary Relationships of Six Axenic Non-Marine Cyanobacteria. Genome Biology and Evolution 11 (1):270-294 |
| Publication: | Cumbers J. et al. (2014) Salt tolerance and polyphyly in the cyanobacterium Chroococcidiopsis (Pleurocapsales). J. Phycol. 50 (3):472–482 |
| Publication: | O'Brien, H.E. et al. (2005) Assessing host specialization in symbiotic cyanobacteria associated with four closely related species of the lichen fungus Peltigera. Eur. J. Phycol. 40 (4):363-378 |
| Publication: | Fewer, D. et al. (2002) Chroococcidiopsis and heterocyst-differentiating cyanobacteria are each other's closest relatives. Mol. Phylogenet. Evol. 23:82-90 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequencing Project: | ASM399189v1 |
| Sequencing Project: | SAMN10102196 |
| Sequence Accession: | JF810050 Chroococcidiopsis cubana SAG 39.79 glutamyl-tRNA synthetase (gltX) gene, partial cds. (483 bp) |
| Sequence Accession: | JF810035 Chroococcidiopsis cubana SAG 39.79 delta-9 acyl-lipid desaturase (desC1) gene, partial cds. (467 bp) |
| Sequence Accession: | JF810065 Chroococcidiopsis cubana SAG 39.79 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds. (978 bp) |
| Sequence Accession: | AJ344558 partial 16S rRNA gene (1422 bp) |
| Sequence Accession: | JF810080 Chroococcidiopsis cubana SAG 39.79 16S ribosomal RNA gene, partial sequence. (1222 bp) |
| Sequence Accession: | GQ980024 Chroococcidiopsis cubana SAG 39.79 23S large subunit ribosomal RNA (rrnL) gene, partial sequence. (630 bp) |
| Sequence Accession: | GCA_003991895 (0 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | no |