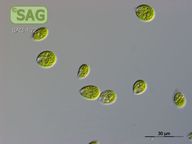
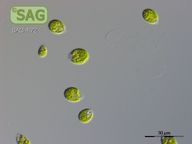
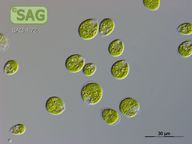
SAG 4.72 Chlamydomonas debaryana
| SAG Strain Number: | 4.72 |
| TAXONOMY | |
| Genus: | Chlamydomonas |
| Species: | debaryana |
| Taxonomic position: | Chlorophyta - Chlorophyceae |
| Authority: | Goroschankin |
| Variety: | |
| Formerly called: | C. angulosa |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN | |
| General habitat: | freshwater |
| Climatic zone: | |
| Continent: | Asia |
| Country: | Japan |
| Locality: | Kobe |
| Lat. / Long. (Precision): | 34.689404 / 135.192204 (3000m) View on Google Maps |
| Year: | before 1955 |
| Isolated by: | Y. Tsubo |
| Strain number by isolator: | |
| Deposition by: | R.C. Starr (UTEX) (< R.A. Lewin 1954) |
| Deposition date: | 1971 |
| CULTURE INFORMATION | |
| Culture medium: | K Ag |
| Axenic: | axenic |
| Strain relatives: | CCAP 11/59; UTEX 618 |
| Agitation resistance: | yes |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; |
| PUBLICATIONS | |
| Publication: | Pröschold T. et al. (2018) Chlamydomonas schloesseri sp. nov. (Chlamydophyceae, Chlorophyta) revealed by morphology, autolysin cross experiments, and multiple gene analyses. Phytotaxa 362 (1):21-38 |
| Publication: | Matsuda Y. et al. (1987) Cell walls of algae in the volvocales: Their sensitivity to a cell wall lytic enzyme and labeling with an anti-cell wall glycopeptide of Chlamydomonas reinhardtii. Bot. Mag. Tokyo 100 (4):373-384 |
| SEQUENCE INFORMATION | |
| Sequence Accession: | MG650081 Chlamydomonas debaryana SAG 4.72 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. (1128 bp) |
| Sequence Accession: | MF677995 Chlamydomonas debaryana strain SAG 4.72 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1 and 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2, partial sequence. (2416 bp) |
| CRYO | |
| Cryopreservation: | not detected |
| TEACHING | |
| Recommended for teaching: | no |