| SAG Strain Number: | 45-1 |
| TAXONOMY |
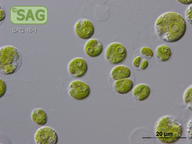
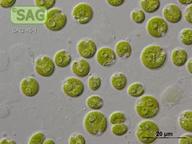
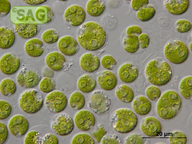
| Genus: | Lobomonas |
| Species: | francei |
| Taxonomic position: | Chlorophyta - Chlorophyceae |
| Authority: | Dangeard emend. Sausen, Malavasi et Melkonian |
| Variety: | |
| Formerly called: | authentic strain of Lobomonas rostrata Hazen; authentic strain of Lobomonas piriformis Pringsheim |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN |
| General habitat: | freshwater |
| Climatic zone: | temperate |
| Continent: | Europe |
| Country: | Czech Republic |
| Locality: | sandy shore of River Elbe near Celakovice |
| Lat. / Long. (Precision): | 50.17437 / 14.726214 (800m) View on Google Maps |
| Year: | 1930 |
| Isolated by: | E.G. Pringsheim |
| Strain number by isolator: | 45-1 |
| Deposition by: | E.G. Pringsheim |
| Deposition date: | 1954 |
| CULTURE INFORMATION |
| Culture medium: | ESP Ag |
| Axenic: | axenic |
| Strain relatives: | CCAP 45/2; UTEX 17; ATCC 30403 |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; OneKP Sample ID: JKKI; authentic strain of Lobomonas rostrata Hazen assigned as non-authentic strain of L. francei Dangeard emend. Sausen, Malavasi et Melkonian 2018. SAG 08/2022 |
| |
| PUBLICATIONS |
| Publication: | Puginier C. et al. (2024) Phylogenomics reveals the evolutionary origins of lichenization in chlorophyte algae. Nat. Comm. 15 (1):4452 |
| Publication: | Goh F.Q.Y. et al. (2019) Gains and losses of metabolic function inferred from a phylotranscriptomic analysis of algae. Sci. Rep. 9:10482 |
| Publication: | Nakada T. et al. (2019) Improved taxon sampling and multigene phylogeny of unicellular chlamydomonads closely related to the colonial volvocalean lineage Tetrabaenaceae-Goniaceae-Volvocaceae (Volvocales, Chlorophyceae). Mol. Phylogen. Evol. 130:1-8 |
| Publication: | Sausen N. et al. (2018) Molecular phylogeny, systematics, and revision of the type species of Lobomonas, L. francei (Volvocales, Chlorophyta) and closely related taxa . J. Phycol. 54 (2):198-214 |
| Publication: | Nakada T. et al. (2016) 18S Ribosomal RNA Gene Phylogeny of a Colonial Volvocalean Lineage (Tetrabaenaceae-Goniaceae-Volvocaceae, Volvocales, Chlorophyceae) and Its Close Relatives. J. Jpn. Bot. 91:345-354 |
| Publication: | Matsuda Y. et al. (1987) Cell walls of algae in the volvocales: Their sensitivity to a cell wall lytic enzyme and labeling with an anti-cell wall glycopeptide of Chlamydomonas reinhardtii. Bot. Mag. Tokyo 100 (4):373-384 |
| Publication: | Pringsheim E.G. (1930) Neue Chlamydomonadaceen, welche in Reinkultur gewonnen wurden. Arch. Protistenk. 69:95-102 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequence Accession: | LC380289 Lobomonas francei SAG 45-1 chloroplast atpB gene for ATP synthase beta-subunit, partial cds. (1207 bp) |
| Sequence Accession: | LC380395 Lobomonas francei SAG 45-1 chloroplast rbcL gene for ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, partial cds. (1139 bp) |
| Sequence Accession: | LC380370 Lobomonas francei SAG 45-1 chloroplast psbC gene for photosystem II CP43 apoprotein, partial cds. (1549 bp) |
| Sequence Accession: | LC380343 Lobomonas francei SAG 45-1 chloroplast psaB gene for photosystem I P700 chlorophyll a apoprotein A2, partial cds. (1516 bp) |
| Sequence Accession: | LC380316 Lobomonas francei SAG 45-1 chloroplast psaA gene for photosystem I P700 chlorophyll a apoprotein A1, partial cds. (1522 bp) |
| Sequence Accession: | LT853996 Lobomonas francei genomic DNA sequence contains 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2 and 28S rRNA gene, strain Lobomonas rostrata. (2901 bp) |
| Sequence Accession: | LC086369 Lobomonas rostrata gene for 18S rRNA, partial sequence, strain: SAG 45-1. (1742 bp) |
| Sequence Accession: | ERS1830112 1000 Plants (1kP) RNA-Seq Transcriptome Sample ID: JKKI , multiple cells from a single clone (0 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | no |