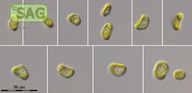
| SAG Strain Number: | 50-1 |
| TAXONOMY |
| Genus: | Mesostigma |
| Species: | viride |
| Taxonomic position: | Streptophyta - Mesostigmatophyceae |
| Authority: | Lauterborn |
| Variety: | |
| Formerly called: | |
| Authentic: | no |
| Division: | Streptophyta |
| Class: | Mesostigmatophyceae |
| ORIGIN |
| General habitat: | freshwater |
| Climatic zone: | |
| Continent: | Europe |
| Country: | United Kingdom |
| Locality: | Cambridge |
| Lat. / Long. (Precision): | 52.202347 / 0.128345 (6000m) View on Google Maps |
| Year: | 1943 |
| Isolated by: | E.G. Pringsheim |
| Strain number by isolator: | |
| Deposition by: | E.G. Pringsheim |
| Deposition date: | 1954 |
| CULTURE INFORMATION |
| Culture medium: | MiEB12 |
| Axenic: | bacterial or other types of contamination |
| Strain relatives: | CCAP 50/1 |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; |
| |
| PUBLICATIONS |
| Publication: | Glass S.E. et al. (2023) Chloroplast genome evolution and phylogeny of the early-diverging charophycean green algae with a focus on the Klebsormidiophyceae and Streptofilum. J. Phycol. 59:1133-1146 |
| Publication: | Goh F.Q.Y. et al. (2019) Gains and losses of metabolic function inferred from a phylotranscriptomic analysis of algae. Sci. Rep. 9:10482 |
| Publication: | Yang, Ashley et al. (2016) Rickettsial endosymbiont in the “early-diverging” streptophyte green alga Mesostigma viride. Journal of Phycology 52 (2 ): 219-229 |
| Publication: | Hua J. et al. (2012) Similar Relative Mutation Rates in the Three Genetic Compartments of Mesostigma and Chlamydomonas. Protist 163:110-115 |
| Publication: | Cocquyt, E. et al. (2010) Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol Biol Evol 27 (9):2052-61 |
| Publication: | Marin B. et al. (2010) Molecular phylogeny and classification of the Mamiellophyceae class. nov. (Chlorophyta) based on sequence comparisons of the nuclear- and plastidencoded rRNA operons. 161 (2):304-336 |
| Publication: | Hall J.D. et al. (2008) PHYLOGENY OF THE CONJUGATING GREEN ALGAE BASED ON CHLOROPLAST AND MITOCHONDRIAL NUCLEOTIDE SEQUENCE DATA. J. Phycol. 44 (2):467-477 |
| Publication: | Karol K.G. et al. (2001) The Closest Living Relatives of Land Plants. Science 294:2351-2353 |
| Publication: | Marin, B. et al. (1999) Mesostigmatophyceae, a New Class of Streptophyte Green Algae Revealed by SSU rRNA Sequence Comparisons. Protist 150:399-417 |
| Publication: | Bhattacharya D. et al. (1998) Actin phylogeny identifies Mesostigma viride as a flagellate ancestor of the land plants. Journal of Molecular Evolution 47 (5):544-550 |
| Publication: | Fawley M.W. (1991) DISJUNCT DISTRIBUTION OF THE XANTHOPHYLL LOROXANTHIN IN THE GREEN ALGAE (CHLOROPHYTA). J. Phycol. 27:544-548 |
| Publication: | Reize I.B. et al. (1989) A New Way to Investigate Living Flagellated/Ciliated Cells in the Light Microscope: Immobilization of Cells in Agarose. Bot. Acta 102 (2):145-151 |
| Publication: | Schulze D. et al. (1987) Immunolocalization of a Ca2+ -modulated contractile protein in the flagellar apparatus of green algae: the nucleus-basal body connector. Eur J Cell Biol 45:51-61 |
| Publication: | McFadden, G.I. et al. (1986) Use of Hepes buffer for microalgal culture media and fixation for electron microscopy. Phycologia 25 (4):551-557 |
| show more publications ... |
| SEQUENCE INFORMATION |
| Sequence Accession: | OR168113 Mesostigma viride strain SAG 50-1 plastid, complete genome. (120172 bp) |
| Sequence Accession: | HQ667981 Mesostigma viride strain SAG 50-1 mitochondrion, partial genome. (45885 bp) |
| Sequence Accession: | HQ668006 Mesostigma viride culture SAG:50-1 strain SAG 50-1 chloroplast sedoheptulose-1, 7-bisphosphatase genes, partial cds; nuclear genes for chloroplast products. (1151 bp) |
| Sequence Accession: | HQ668004 Mesostigma viride culture SAG:50-1 strain SAG 50-1 glyceraldehyde-3-phosphate dehydrogenase gene, partial cds. (1126 bp) |
| Sequence Accession: | HQ668002 Mesostigma viride culture SAG:50-1 strain SAG 50-1 14-3-3-like protein gene, partial cds. (912 bp) |
| Sequence Accession: | HQ668000 Mesostigma viride culture SAG:50-1 strain SAG 50-1 ubiquitin extension protein gene, partial cds. (977 bp) |
| Sequence Accession: | HQ667998 Mesostigma viride culture SAG:50-1 strain SAG 50-1 actin gene, partial cds. (1179 bp) |
| Sequence Accession: | AF061020 Mesostigma viride strain SAG 50-1 culture-collection SAG:50-1 actin gene, complete cds. (1504 bp) |
| Sequence Accession: | AF408222 Mesostigma viride strain SAG 50-1 voucher SAG:50-1 NADH dehydrogenase subunit 5 (nad5) gene, partial cds; mitochondrial. (783 bp) |
| Sequence Accession: | AF408245 Mesostigma viride strain SAG 50-1 voucher SAG:50-1 18S small subunit ribosomal RNA gene, partial sequence. (1652 bp) |
| Sequence Accession: | AF408256 Mesostigma viride strain SAG 50-1 voucher SAG:50-1 ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. (1353 bp) |
| Sequence Accession: | AF408806 Mesostigma viride strain SAG 50-1 voucher SAG:50-1 ATP synthase beta subunit (atpB) gene, partial cds; chloroplast. (1206 bp) |
| Sequence Accession: | AY591912 Mesostigma viride strain SAG 50-1 26S ribosomal RNA gene, partial sequence. (1373 bp) |
| Sequence Accession: | KJ395942 Mesostigma viride ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene, partial cds; chloroplast. (1354 bp) |
| Sequence Accession: | KJ395895 Mesostigma viride CP43 chlorophyll apoprotein of photosystem II (psbC) gene, partial cds; chloroplast. (1194 bp) |
| Sequence Accession: | AJ250108 Mesostigma viride partial 18S rRNA gene, strain SAG 50-1. (1756 bp) |
| Sequence Accession: | FN563105 Mesostigma viride plastid partial 16S rRNA gene, strain SAG 50-1. (1459 bp) |
| Sequence Accession: | EF371171 Mesostigma viride strain SAG 501 cytochrome oxidase subunit III (cox III) gene, partial cds; mitochondrial. (609 bp) |
| Sequence Accession: | EF371278 Mesostigma viride strain SAG 501 photosystem I P700 chlorophyll A apoprotein A1 (psaA) gene, partial cds; chloroplast. (2044 bp) |
| show more sequences ... |
| CRYO |
| Cryopreservation: | not detected |
| TEACHING |
| Recommended for teaching: | yes |