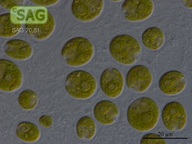
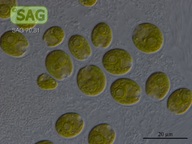
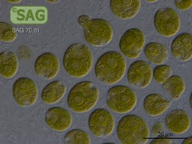
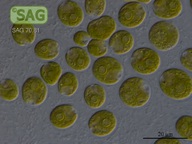
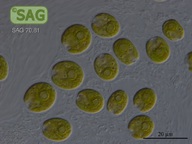
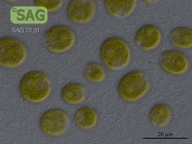
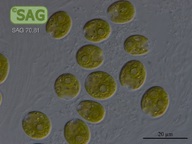
SAG 70.81 Chlamydomonas latifrons cf.
| SAG Strain Number: | 70.81 |
| TAXONOMY | |
| Genus: | Chlamydomonas |
| Species: | latifrons cf. |
| Taxonomic position: | Chlorophyta - Chlorophyceae |
| Authority: | Nygaard |
| Variety: | |
| Formerly called: | Chlamydomonas debaryana Goroschankin |
| Authentic: | no |
| Division: | Chlorophyta |
| Class: | Chlorophyceae |
| Common name: | Green Algae - Chlorophytes |
| ORIGIN | |
| General habitat: | |
| Climatic zone: | temperate |
| Continent: | Europe |
| Country: | Czech Republic |
| Locality: | Brno |
| Lat. / Long. (Precision): | |
| Year: | before 1973 |
| Isolated by: | H. Ettl |
| Strain number by isolator: | as Chlamydomonas inepta var. lateovalis Ettl |
| Deposition by: | H. Ettl |
| Deposition date: | 1973 |
| CULTURE INFORMATION | |
| Culture medium: | ES |
| Axenic: | bacterial and fungal contaminations |
| Strain relatives: | |
| Agitation resistance: | not detected |
| Special properties: | NO known Nagoya Protocol restrictions for this strain; acc. to Proeschold et al. 2018 not a C. debaryana Goroschankin but C. cf. latifrons until further examinations. SAG 08/2022 |
| PUBLICATIONS | |
| Publication: | Lindsey C.R. et al. (2021) Phylotranscriptomics points to multiple independent origins of multicellularity and cellular differentiation in the volvocine algae. BMC Biol. 19:182 |
| Publication: | Pröschold T. et al. (2018) Chlamydomonas schloesseri sp. nov. (Chlamydophyceae, Chlorophyta) revealed by morphology, autolysin cross experiments, and multiple gene analyses. Phytotaxa 362 (1):21-38 |
| SEQUENCE INFORMATION | |
| Sequencing Project: | SAMN17884633 BioSample: SAMN17884633; Sample name: Chlamydomonas debaryana S 70.81; SRA: SRS8264728 |
| Sequence Accession: | MF678038 Chlamydomonas cf. latifrons SAG 70.81 small subunit ribosomal RNA gene, partial sequence; internal transcribed spacer 1 and 5.8S ribosomal RNA gene, complete sequence; and internal transcribed spacer 2, partial sequence. (2407 bp) |
| CRYO | |
| Cryopreservation: | not detected |
| TEACHING | |
| Recommended for teaching: | no |